Classes that inherit from the QMatrix class handle everything associated with the instantaneous rate matrix. The Model simply has to send the exchangeabilities and state frequencies to its data member _qmatrix, and the QMatrix objects stored there handle computing the eigenvalues, eigenvectors, and inverse eigenvectors that BeagleLib needs.
This tutorial emplements two different kinds of models: (1) the GTR+I+G model for nucleotide data and (2) a codon model. These two models have rate matrices that are different size (4x4 for GTR and 61x61 for a codon model using the standard code) and, rather than create one generic class to handle both, we will create an abstract base class QMatrix and two derived classes, QMatrixNucleotide (for the GTR case) and QMatrixCodon (for the codon case).
Begin by creating a new header file named qmatrix.hpp and add the QMatrix class declaration and member function bodies:
#pragma once
#include <algorithm>
#include <vector>
#include <Eigen/Dense>
#include "genetic_code.hpp"
#include "xstrom.hpp"
namespace strom {
class QMatrix {
public:
typedef std::vector<double> freq_xchg_t;
typedef std::shared_ptr<freq_xchg_t> freq_xchg_ptr_t;
typedef double omega_t;
typedef std::shared_ptr<omega_t> omega_ptr_t;
typedef boost::shared_ptr<QMatrix> SharedPtr;
QMatrix();
virtual ~QMatrix();
virtual void clear() = 0;
virtual void setEqualStateFreqs(freq_xchg_ptr_t freq_ptr) = 0;
virtual void setStateFreqsSharedPtr(freq_xchg_ptr_t freq_ptr) = 0;
virtual void setStateFreqs(freq_xchg_t & freq) = 0;
virtual freq_xchg_ptr_t getStateFreqsSharedPtr() = 0;
virtual const double * getStateFreqs() const = 0;
void fixStateFreqs(bool is_fixed);
bool isFixedStateFreqs() const;
virtual void setEqualExchangeabilities(freq_xchg_ptr_t xchg_ptr) = 0;
virtual void setExchangeabilitiesSharedPtr(freq_xchg_ptr_t xchg) = 0;
virtual void setExchangeabilities(freq_xchg_t & xchg) = 0;
virtual freq_xchg_ptr_t getExchangeabilitiesSharedPtr() = 0;
virtual const double * getExchangeabilities() const = 0;
void fixExchangeabilities(bool is_fixed);
bool isFixedExchangeabilities() const;
virtual void setOmegaSharedPtr(omega_ptr_t omega) = 0;
virtual void setOmega(omega_t omega) = 0;
virtual omega_ptr_t getOmegaSharedPtr() = 0;
virtual double getOmega() const = 0;
void fixOmega(bool is_fixed);
bool isFixedOmega() const;
virtual const double * getEigenvectors() const = 0;
virtual const double * getInverseEigenvectors() const = 0;
virtual const double * getEigenvalues() const = 0;
void setActive(bool activate);
protected:
virtual void recalcRateMatrix() = 0;
void normalizeFreqsOrExchangeabilities(freq_xchg_ptr_t v);
bool _is_active;
bool _state_freqs_fixed;
bool _exchangeabilities_fixed;
bool _omega_fixed;
};
// member function bodies go here
// QMatrixNucleotide class goes here
// QMatrixCodon class goes here
}
That QMatrix is an abstract class is clear from the fact that most member functions are set equal to 0 in the class declaration and no body is provided for them. Being abstract, this class cannot be used to create objects; we must derive other classes from it that provide bodies for the member functions.
The setActive, clear, fixStateFreqs, fixExchangeabilities, fixOmega, and normalizeFreqsOrExchangeabilities member functions are defined (i.e. have a body), which means that inherited classes will have their functionality also.
Constructor and destructor
inline QMatrix::QMatrix() {
//std::cout << "Creating a QMatrix object" << std::endl;
}
inline QMatrix::~QMatrix() {
//std::cout << "Destroying a QMatrix object" << std::endl;
}
The setActive member function
If setActive is passed false, then this QMatrix (or QMatrix-derived) object becomes inactive, meaning it will accept new state frequencies, exchangeabilities, or omega values but will not calculate anything with those values. This will be useful later when we run an MCMC analysis without data, as it allows us to avoid expensive eigensystem computations when the likelihood is not even being calculated. The boolean value passed into this function is assigned to the _is_active data member, and, if _is_active has just become true, it calls recalcRateMatrix just in case the eigenvalues and eigenvectors are not up-to-date. Because recalcRateMatrix is a virtual function, the appropriate implementation of recalcRateMatrix will be used (QMatrixNucleotide::recalcRateMatrix vs. QMatrixCodon::recalcRateMatrix).
inline void QMatrix::setActive(bool activate) {
_is_active = activate;
recalcRateMatrix();
}
The clear member function
This function provides initial values for the four data members (_is_active, _state_freqs_fixed, _exchangeabilities_fixed, and _omega_fixed) that are defined in this base class.
inline void QMatrix::clear() {
_is_active = false;
_state_freqs_fixed = false;
_exchangeabilities_fixed = false;
_omega_fixed = false;
}
The fixStateFreqs, fixExchangeabilities, and fixOmega member functions
These three functions provide a way to change the values of the _state_freqs_fixed, _exchangeabilities_fixed, and _omega_fixed data members, respectively.
inline void QMatrix::fixStateFreqs(bool is_fixed) {
_state_freqs_fixed = is_fixed;
}
inline void QMatrix::fixExchangeabilities(bool is_fixed) {
_exchangeabilities_fixed = is_fixed;
}
inline void QMatrix::fixOmega(bool is_fixed) {
_omega_fixed = is_fixed;
}
The isFixedStateFreqs, isFixedExchangeabilities, and isFixedOmega member functions
These three functions are accessors for the values of the _state_freqs_fixed, _exchangeabilities_fixed, and _omega_fixed data members, respectively.
inline bool QMatrix::isFixedStateFreqs() const {
return _state_freqs_fixed;
}
inline bool QMatrix::isFixedExchangeabilities() const {
return _exchangeabilities_fixed;
}
inline bool QMatrix::isFixedOmega() const {
return _omega_fixed;
}
The normalizeFreqsOrExchangeabilities member function
This function takes a shared pointer to either a state frequencies vector or an exhangeabilities vector and normalizes the vector so that its elements sum to 1.0.
inline void QMatrix::normalizeFreqsOrExchangeabilities(QMatrix::freq_xchg_ptr_t v) {
// Be sure elements of v sum to 1.0 and assert that they are all positive
double sum_v = std::accumulate(v->begin(), v->end(), 0.0);
for (auto & x : *v) {
assert(x > 0.0);
x /= sum_v;
}
}
The QMatrixNucleotide class
We will keep all three classes in this same qmatrix.hpp file, so go ahead and add the declaration for the QMatrixNucleotide class after the body of QMatrix::setActive but before the right curly bracket that closes the strom namespace.
class QMatrixNucleotide : public QMatrix {
public:
typedef Eigen::Matrix<double, 4, 4, Eigen::RowMajor> eigenMatrix4d_t;
typedef Eigen::Vector4d eigenVector4d_t;
QMatrixNucleotide();
~QMatrixNucleotide();
void clear();
void setEqualStateFreqs(freq_xchg_ptr_t freq_ptr);
void setStateFreqsSharedPtr(freq_xchg_ptr_t freq_ptr);
void setStateFreqs(freq_xchg_t & freqs);
freq_xchg_ptr_t getStateFreqsSharedPtr();
const double * getStateFreqs() const;
void setEqualExchangeabilities(freq_xchg_ptr_t xchg_ptr);
void setExchangeabilitiesSharedPtr(freq_xchg_ptr_t xchg_ptr);
void setExchangeabilities(freq_xchg_t & xchg);
freq_xchg_ptr_t getExchangeabilitiesSharedPtr();
const double * getExchangeabilities() const;
void setOmegaSharedPtr(omega_ptr_t omega_ptr);
void setOmega(omega_t omega);
omega_ptr_t getOmegaSharedPtr();
double getOmega() const;
const double * getEigenvectors() const;
const double * getInverseEigenvectors() const;
const double * getEigenvalues() const;
EIGEN_MAKE_ALIGNED_OPERATOR_NEW
protected:
virtual void recalcRateMatrix();
private:
// workspaces for computing eigenvectors/eigenvalues
eigenMatrix4d_t _sqrtPi;
eigenMatrix4d_t _sqrtPiInv;
eigenMatrix4d_t _Q;
eigenMatrix4d_t _eigenvectors;
eigenMatrix4d_t _inverse_eigenvectors;
eigenVector4d_t _eigenvalues;
freq_xchg_ptr_t _state_freqs;
freq_xchg_ptr_t _exchangeabilities;
};
// QMatrixNucleotide member function bodies go here
I will provide explanations for the member functions in QMatrixNucleotide, but then just provide the code without explanation for the QMatrixCodon class: the explanations are the same; it is only the actual implementations that differ.
The Eigen library is used to do the work of computing eigenvalues and eigenvectors, which explains the #include <Eigen/Dense> at the top of the file. The typedefs eigenMatrix4d_t and eigenVector4d_t simplify specify matrices and vectors, respectively, that are used with the Eigen library. You’ll note that these types specify vectors of length 4 and 4 by 4 matrices, so we are clearly designing our QMatrixNucleotide class to handle only 4-state nucleotide data.
The constructor, destructor, and clear functions
The constructor and destructor are trivial, as usual. The clear function sets up the parameters _exchangeabilities and _state_freqs according to the JC model. Model parameters are stored as shared pointers so that they can be easily linked across data subsets.
inline QMatrixNucleotide::QMatrixNucleotide() {
//std::cout << "Constructing a QMatrixNucleotide object" << std::endl;
clear();
}
inline QMatrixNucleotide::~QMatrixNucleotide() {
//std::cout << "Destroying a QMatrixNucleotide object" << std::endl;
}
inline void QMatrixNucleotide::clear() {
QMatrix::clear();
QMatrix::freq_xchg_t xchg = {1,1,1,1,1,1};
_exchangeabilities = std::make_shared<QMatrix::freq_xchg_t>(xchg);
QMatrix::freq_xchg_t freq_vect = {0.25, 0.25, 0.25, 0.25};
_state_freqs = std::make_shared<QMatrix::freq_xchg_t>(freq_vect);
recalcRateMatrix();
}
The getExchangeabilitiesSharedPtr and getStateFreqsSharedPtr member functions
These two functions are accessors for _exchangeabilities and _state_freqs, respectively.
inline QMatrix::freq_xchg_ptr_t QMatrixNucleotide::getExchangeabilitiesSharedPtr() {
return _exchangeabilities;
}
inline QMatrix::freq_xchg_ptr_t QMatrixNucleotide::getStateFreqsSharedPtr() {
return _state_freqs;
}
The getOmegaSharedPtr member function
You may be thinking that a member function named getOmegaSharedPtr makes no sense in the QMatrixNucleotide class. You are correct, but we must implement it anyway, otherwise QMatrixNucleotide would be an abstract class like QMatrix and we would not be able to create objects from it. Because it makes no sense to call this function, we use an assert(false) to let us know if this function is ever used and return nullptr to avoid a compiler error (we must return a pointer, even if it points to nothing!).
inline QMatrix::omega_ptr_t QMatrixNucleotide::getOmegaSharedPtr() {
assert(false);
return nullptr;
}
The getEigenvectors, getInverseEigenvectors, and getEigenvalues member functions
These are accessors that return pointers to the C-style arrays beneath the hood of the Eigen::Matrix data members _eigenvectors, _inverse_eigenvectors, and _eigenvalues, respectively. The reason we do not return references to the Eigen::Matrix objects directly is that these functions are used to fill buffers in BeagleLib, which expects C-style arrays.
inline const double * QMatrixNucleotide::getEigenvectors() const {
return _eigenvectors.data();
}
inline const double * QMatrixNucleotide::getInverseEigenvectors() const {
return _inverse_eigenvectors.data();
}
inline const double * QMatrixNucleotide::getEigenvalues() const {
return _eigenvalues.data();
}
The getExchangeabilities and getStateFreqs member functions
These two functions return pointers to the C-style arrays beneath the hood of the data members _exchangeabilities and _state_freqs, respectively. The reason we do not return the shared pointers _exchangeabilities and _state_freqs themselves is that these functions are used to fill buffers in BeagleLib, which expects C-style arrays. In order to obtain the C-style array from a std::shared_ptr pointing to a std::vector, first obtain a reference to the vector pointed to (*), then obtain the memory address (&) of the first element of the vector ([0]).
inline const double * QMatrixNucleotide::getExchangeabilities() const {
return &(*_exchangeabilities)[0];
}
inline const double * QMatrixNucleotide::getStateFreqs() const {
return &(*_state_freqs)[0];
}
The getOmega member function
This function should not be called, as the QMatrixNucleotide class does not use the omega parameter, so the function uses assert(false) to catch any usage in debug mode and returns 0.0.
inline double QMatrixNucleotide::getOmega() const {
assert(false);
return 0.0;
}
The setEqualExchangeabilities member function
This function sets all 6 exchangeabilities to 1/6 and calls the recalcRateMatrix function before returning.
inline void QMatrixNucleotide::setEqualExchangeabilities(QMatrix::freq_xchg_ptr_t xchg_ptr) {
_exchangeabilities = xchg_ptr;
_exchangeabilities->assign(6, 1.0/6.0);
recalcRateMatrix();
}
The setExchangeabilitiesSharedPtr member function
This function copies the supplied shared pointer to a vector of exchangeabilities (xchg_ptr) to the data member _exchangeabilities (after checking to make sure it is the correct length), then calls the recalcRateMatrix function before returning.
inline void QMatrixNucleotide::setExchangeabilitiesSharedPtr(QMatrix::freq_xchg_ptr_t xchg_ptr) {
if (xchg_ptr->size() != 6)
throw XStrom(boost::format("Expecting 6 exchangeabilities and got %d: perhaps you meant to specify a subset data type other than nucleotide") % xchg_ptr->size());
_exchangeabilities = xchg_ptr;
normalizeFreqsOrExchangeabilities(_exchangeabilities);
recalcRateMatrix();
}
The setExchangeabilities member function
This function copies the supplied vector of exchangeabilities (xchg) to the vector pointed to by the data member _exchangeabilities (after checking to make sure it is the correct length), then calls the recalcRateMatrix function before returning.
inline void QMatrixNucleotide::setExchangeabilities(QMatrix::freq_xchg_t & xchg) {
if (xchg.size() != 6)
throw XStrom(boost::format("Expecting 6 exchangeabilities and got %d: perhaps you meant to specify a subset data type other than nucleotide") % xchg.size());
std::copy(xchg.begin(), xchg.end(), _exchangeabilities->begin());
recalcRateMatrix();
}
The setEqualStateFreqs member function
This function sets all 4 state frequencies to 0.25 and calls the recalcRateMatrix function before returning.
inline void QMatrixNucleotide::setEqualStateFreqs(QMatrix::freq_xchg_ptr_t freq_ptr) {
_state_freqs = freq_ptr;
_state_freqs->assign(4, 0.25);
recalcRateMatrix();
}
The setStateFreqsSharedPtr member function
This function copies the supplied shared pointer to a vector of state frequencies (freq_ptr) to the data member _state_freqs (after checking to make sure it is the correct length). It then calls the recalcRateMatrix function before returning.
inline void QMatrixNucleotide::setStateFreqsSharedPtr(QMatrix::freq_xchg_ptr_t freq_ptr) {
if (freq_ptr->size() != 4)
throw XStrom(boost::format("Expecting 4 state frequencies and got %d: perhaps you meant to specify a subset data type other than nucleotide") % freq_ptr->size());
double sum_of_freqs = std::accumulate(freq_ptr->begin(), freq_ptr->end(), 0.0);
if (std::fabs(sum_of_freqs - 1.0) > 0.001)
throw XStrom(boost::format("Expecting sum of 4 state frequencies to be 1, but instead got %g") % sum_of_freqs);
_state_freqs = freq_ptr;
recalcRateMatrix();
}
The setStateFreqs member function
This function copies the supplied vector of state frequencies (freqs) to the vector pointed to by the data member _state_freqs (after checking to make sure it is the correct length). It then calls the recalcRateMatrix function before returning.
inline void QMatrixNucleotide::setStateFreqs(QMatrix::freq_xchg_t & freqs) {
if (freqs.size() != 4)
throw XStrom(boost::format("Expecting 4 state frequencies and got %d: perhaps you meant to specify a subset data type other than nucleotide") % freqs.size());
std::copy(freqs.begin(), freqs.end(), _state_freqs->begin());
recalcRateMatrix();
}
The setOmegaSharedPtr amd setOmega member functions
Because the omega parameter is not used in the GTR model, these two functions should not be called at all, which explains why their only line is assert(false) to catch any usage of these functions in the debug version of the program.
inline void QMatrixNucleotide::setOmegaSharedPtr(QMatrix::omega_ptr_t omega_ptr) {
assert(false);
}
inline void QMatrixNucleotide::setOmega(QMatrix::omega_t omega) {
assert(false);
}
The recalcRateMatrix member function
This function requires more explanation than any other, and I’ve broken down the discussion into sections.
GTR rate matrix
The instantaneous rate matrix Q for the GTR model has 6 exchangeability parameters (a, b, c, d, e, and f) and 4 nucleotide frequencies (πA, πC, πG and πT). It is stored in the data member _qmatrix.
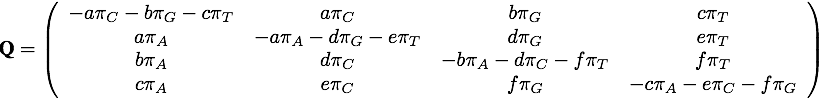
The rows represent the “from” state (in the order A, C, G, T, from top to bottom), while the columns represent the “to” state (also in the order A, C, G, T, from left to right). The diagonal elements of this matrix are negative and equal to the sum of the other elements in the same row because any change from state A, for example, to a different state must reduce the amount of A, hence the rate of change is negative. The row sums are zero because this model assumes that the sequence length neither shrinks nor grows, and thus any increase in C, G, or T must occur at the expense of A.
Matrices related to nucleotide frequencies
Two important diagonal matrices are defined by the vector of nucleotide frequencies stored in _state_freqs. These are shown below and are stored in the data members _sqrtPi (top) and _sqrtPiInv (bottom).
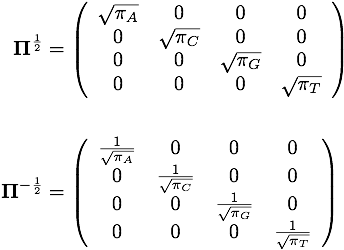
Diagonalization of the rate matrix
The rate matrix Q can be represented as the matrix product of the (right) eigenvector matrix V, the diagonal matrix of eigenvalues L, and the inverse eigenvector matrix V-1.

Computing the transition probability matrix
The transition probability matrix P is obtained by exponentiating the product of Q and time t. The matrix Q is scaled so that time t can be measured in units of expected substitutions per site, which is why the rates in the Q matrix omit the overall rate of substitution and only include relative rate terms (the overall rate is subsumed in t). The calculation of P involves exponentiating only the middle matrix (diagonal matrix of eigenvalues multiplied by t).
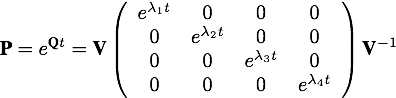
Scaling the rate matrix
The Q matrix must be scaled so that time t is measured in expected number of substitutions per site. If this scaling is not performed, the edge lengths lose their interpretation as expected number of substitutions per site. The scaling factor needed may be obtained by effectively performing the following matrix multiplication and afterwards summing the off-diagonal elements: dividing each element of Q by this sum yields the desired normalization.
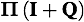
Symmetrizing the rate matrix
Computing eigenvalues and eigenvectors of a symmetric matrix is simpler and more efficient than computing them for an asymmetric matrix. Hence, it is common to obtain eigenvalues and eigenvectors of a symmetrical matrix S derived from the Q matrix rather than for the Q matrix itself. The eigenvalues for S and Q are identical, and the eigenvector matrix W obtained from S may be easily converted into the desired matrix V containing the eigenvectors of Q. Note that V may be obtained by premultiplying W by the diagonal matrix of inverse square roots of nucleotide frequencies.
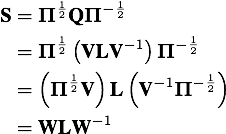
The recalcRateMatrix function
With all that in mind, hopefully the recalcRateMatrix function will be easier to understand.
inline void QMatrixNucleotide::recalcRateMatrix() {
// Must have assigned both _state_freqs and _exchangeabilities to recalculate rate matrix
if (!_is_active || !(_state_freqs && _exchangeabilities))
return;
double piA = (*_state_freqs)[0];
double piC = (*_state_freqs)[1];
double piG = (*_state_freqs)[2];
double piT = (*_state_freqs)[3];
Eigen::Map<const Eigen::Array4d> tmp(_state_freqs->data());
_sqrtPi = tmp.sqrt().matrix().asDiagonal();
_sqrtPiInv = _sqrtPi.inverse();
assert(_exchangeabilities->size() == 6);
double rAC = (*_exchangeabilities)[0];
double rAG = (*_exchangeabilities)[1];
double rAT = (*_exchangeabilities)[2];
double rCG = (*_exchangeabilities)[3];
double rCT = (*_exchangeabilities)[4];
double rGT = (*_exchangeabilities)[5];
double inverse_scaling_factor = piA*(rAC*piC + rAG*piG + rAT*piT) + piC*(rAC*piA + rCG*piG + rCT*piT) + piG*(rAG*piA + rCG*piC + rGT*piT) + piT*(rAT*piA + rCT*piC + rGT*piG);
double scaling_factor = 1.0/inverse_scaling_factor;
_Q(0,0) = -scaling_factor*(rAC*piC + rAG*piG + rAT*piT);
_Q(0,1) = scaling_factor*rAC*piC;
_Q(0,2) = scaling_factor*rAG*piG;
_Q(0,3) = scaling_factor*rAT*piT;
_Q(1,0) = scaling_factor*rAC*piA;
_Q(1,1) = -scaling_factor*(rAC*piA + rCG*piG + rCT*piT);
_Q(1,2) = scaling_factor*rCG*piG;
_Q(1,3) = scaling_factor*rCT*piT;
_Q(2,0) = scaling_factor*rAG*piA;
_Q(2,1) = scaling_factor*rCG*piC;
_Q(2,2) = -scaling_factor*(rAG*piA + rCG*piC + rGT*piT);
_Q(2,3) = scaling_factor*rGT*piT;
_Q(3,0) = scaling_factor*rAT*piA;
_Q(3,1) = scaling_factor*rCT*piC;
_Q(3,2) = scaling_factor*rGT*piG;
_Q(3,3) = -scaling_factor*(rAT*piA + rCT*piC + rGT*piG);
// S is a symmetric matrix
eigenMatrix4d_t S = eigenMatrix4d_t(_sqrtPi*_Q*_sqrtPiInv);
// Can use efficient eigensystem solver because S is symmetric
Eigen::SelfAdjointEigenSolver<Eigen::Matrix4d> solver(S);
if (solver.info() != Eigen::Success) {
throw XStrom("Error in the calculation of eigenvectors and eigenvalues of the GTR rate matrix");
}
_eigenvectors = _sqrtPiInv*solver.eigenvectors();
_inverse_eigenvectors = solver.eigenvectors().transpose()*_sqrtPi;
_eigenvalues = solver.eigenvalues();
}
The QMatrixCodon class
The codon model implemented in this tutorial is very simple. It allows rates to differ depending on whether a change is synonymous or nonsynonymous (the omega parameter governs the nonsynonymous/synonymous rate ratio) and it allows codon frequencies to be unequal, which allows for codon usage bias but is a bit costly computationally due to the fact that the state frequency vector has 61 elements (or 60 or 62, depending on the genetic code used). Our codon model does not take transition/transversion bias into account, but that could be fairly easily added.
Here is the QMatrixCodon class in its entirety. Some explanation of how the rate matrix is constructed follows the code; explanations of the QMatrixNucleotide functions above apply equally well to most of the QMatrixCodon functions, with the exception of those member functions pertaining to the omega parameter, which of course are implemented in QMatrixCodon, while in this case it is the exchangeabilities that are not used (and assert(false) statements are used to detect any calls made to functions that get or set exchangeabilities).
Note that we are using Eigen::VectorXd now as the type of _eigenvalues instead of the Eigen::Vector4d that we used in QMatrixNucleotide. The Xd means that the size of the vector is not hard coded, which is important because codon models vary in the number of states depending on which genetic code applies (due to variation in the number of stop codons). Also, the Eigen::Matrix type that is used for _eigenvectors and other matrix data members has Eigen::Dynamic instead of 4 as template arguments. This generality (with its added computational cost) is not required for the GTR model, so the polymorphism of the QMatrix family allows us to use fixed-size matrices and vectors in QMatrixNucleotide to make the eigensystem calculations more efficient for the very common case of nucleotide data and GTR-like models.
class QMatrixCodon : public QMatrix {
public:
typedef Eigen::Matrix<double, Eigen::Dynamic, Eigen::Dynamic, Eigen::RowMajor> eigenMatrixXd_t;
typedef Eigen::VectorXd eigenVectorXd_t;
QMatrixCodon(GeneticCode::SharedPtr gcode);
~QMatrixCodon();
void clear();
void setEqualStateFreqs(freq_xchg_ptr_t freq_ptr);
void setStateFreqsSharedPtr(freq_xchg_ptr_t freq_ptr);
void setStateFreqs(freq_xchg_t & freqs);
freq_xchg_ptr_t getStateFreqsSharedPtr();
const double * getStateFreqs() const;
void setEqualExchangeabilities(freq_xchg_ptr_t xchg_ptr);
void setExchangeabilitiesSharedPtr(freq_xchg_ptr_t xchg_ptr);
void setExchangeabilities(freq_xchg_t & xchg);
freq_xchg_ptr_t getExchangeabilitiesSharedPtr();
const double * getExchangeabilities() const;
void setOmegaSharedPtr(omega_ptr_t omega_ptr);
void setOmega(omega_t omega);
omega_ptr_t getOmegaSharedPtr();
double getOmega() const;
const double * getEigenvectors() const;
const double * getInverseEigenvectors() const;
const double * getEigenvalues() const;
protected:
virtual void recalcRateMatrix();
private:
// workspaces for computing eigenvectors/eigenvalues
eigenMatrixXd_t _sqrtPi;
eigenMatrixXd_t _sqrtPiInv;
eigenMatrixXd_t _Q;
eigenMatrixXd_t _eigenvectors;
eigenMatrixXd_t _inverse_eigenvectors;
eigenVectorXd_t _eigenvalues;
freq_xchg_ptr_t _state_freqs;
omega_ptr_t _omega;
std::vector<std::string> _codons;
std::vector<unsigned> _amino_acids;
GeneticCode::SharedPtr _genetic_code;
};
inline QMatrixCodon::QMatrixCodon(GeneticCode::SharedPtr gcode) {
//std::cout << "Constructing a QMatrixCodon object" << std::endl;
assert(gcode);
_genetic_code = gcode;
clear();
}
inline QMatrixCodon::~QMatrixCodon() {
//std::cout << "Destroying a QMatrixCodon object" << std::endl;
}
inline void QMatrixCodon::clear() {
QMatrix::clear();
unsigned nstates = _genetic_code->getNumNonStopCodons();
_genetic_code->copyCodons(_codons);
_genetic_code->copyAminoAcids(_amino_acids);
QMatrix::omega_t omega = 0.1;
_omega = std::make_shared<QMatrix::omega_t>(omega);
QMatrix::freq_xchg_t freq_vect(nstates, 1./nstates);
_state_freqs = std::make_shared<QMatrix::freq_xchg_t>(freq_vect);
_sqrtPi.resize(nstates, nstates);
_sqrtPiInv.resize(nstates, nstates);
_Q.resize(nstates, nstates);
_eigenvectors.resize(nstates, nstates);
_inverse_eigenvectors.resize(nstates, nstates);
_eigenvalues.resize(nstates);
recalcRateMatrix();
}
inline QMatrix::freq_xchg_ptr_t QMatrixCodon::getExchangeabilitiesSharedPtr() {
assert(false);
return nullptr;
}
inline QMatrix::freq_xchg_ptr_t QMatrixCodon::getStateFreqsSharedPtr() {
return _state_freqs;
}
inline QMatrix::omega_ptr_t QMatrixCodon::getOmegaSharedPtr() {
return _omega;
}
inline const double * QMatrixCodon::getEigenvectors() const {
return _eigenvectors.data();
}
inline const double * QMatrixCodon::getInverseEigenvectors() const {
return _inverse_eigenvectors.data();
}
inline const double * QMatrixCodon::getEigenvalues() const {
return _eigenvalues.data();
}
inline const double * QMatrixCodon::getExchangeabilities() const {
assert(false);
return 0;
}
inline const double * QMatrixCodon::getStateFreqs() const {
return &(*_state_freqs)[0];
}
inline double QMatrixCodon::getOmega() const {
return *_omega;
}
inline void QMatrixCodon::setEqualExchangeabilities(QMatrix::freq_xchg_ptr_t xchg_ptr) {
assert(false);
}
inline void QMatrixCodon::setExchangeabilitiesSharedPtr(QMatrix::freq_xchg_ptr_t xchg_ptr) {
assert(false);
}
inline void QMatrixCodon::setExchangeabilities(QMatrix::freq_xchg_t & xchg) {
assert(false);
}
inline void QMatrixCodon::setEqualStateFreqs(QMatrix::freq_xchg_ptr_t freq_ptr) {
_state_freqs = freq_ptr;
unsigned nstates = _genetic_code->getNumNonStopCodons();
_state_freqs->assign(nstates, 1./nstates);
recalcRateMatrix();
}
inline void QMatrixCodon::setStateFreqsSharedPtr(QMatrix::freq_xchg_ptr_t freq_ptr) {
unsigned nstates = _genetic_code->getNumNonStopCodons();
if (freq_ptr->size() != nstates)
throw XStrom(boost::format("Expecting %d state frequencies and got %d: perhaps you meant to specify a subset data type other than codon") % nstates % freq_ptr->size());
double sum_of_freqs = std::accumulate(freq_ptr->begin(), freq_ptr->end(), 0.0);
if (std::fabs(sum_of_freqs - 1.0) > 0.001)
throw XStrom(boost::format("Expecting sum of codon frequencies to be 1, but instead got %g") % sum_of_freqs);
_state_freqs = freq_ptr;
normalizeFreqsOrExchangeabilities(_state_freqs);
recalcRateMatrix();
}
inline void QMatrixCodon::setStateFreqs(QMatrix::freq_xchg_t & freqs) {
unsigned nstates = _genetic_code->getNumNonStopCodons();
if (freqs.size() != nstates)
throw XStrom(boost::format("Expecting %d state frequencies and got %d: perhaps you meant to specify a subset data type other than codon") % nstates % freqs.size());
std::copy(freqs.begin(), freqs.end(), _state_freqs->begin());
recalcRateMatrix();
}
inline void QMatrixCodon::setOmegaSharedPtr(QMatrix::omega_ptr_t omega_ptr) {
_omega = omega_ptr;
recalcRateMatrix();
}
inline void QMatrixCodon::setOmega(QMatrix::omega_t omega) {
*_omega = omega;
recalcRateMatrix();
}
inline void QMatrixCodon::recalcRateMatrix() {
// Must have assigned both _state_freqs and _omega to recalculate rate matrix
if (!_is_active || !(_state_freqs && _omega))
return;
unsigned nstates = _genetic_code->getNumNonStopCodons();
assert(_state_freqs->size() == nstates);
const double * pi = getStateFreqs();
double omega = getOmega();
Eigen::Map<const Eigen::ArrayXd> tmp(_state_freqs->data(), nstates);
_sqrtPi = tmp.sqrt().matrix().asDiagonal();
_sqrtPiInv = _sqrtPi.inverse();
// Calculate (unscaled) instantaneous rate matrix
_Q = Eigen::MatrixXd::Zero(nstates,nstates);
for (unsigned i = 0; i < nstates-1; i++) {
for (unsigned j = i+1; j < nstates; j++) {
unsigned diffs = 0;
if (_codons[i][0] != _codons[j][0])
diffs++;
if (_codons[i][1] != _codons[j][1])
diffs++;
if (_codons[i][2] != _codons[j][2])
diffs++;
if (diffs == 1) {
bool synonymous = _amino_acids[i] == _amino_acids[j];
_Q(i,j) = (synonymous ? 1.0 : omega)*pi[j];
_Q(j,i) = (synonymous ? 1.0 : omega)*pi[i];
_Q(i,i) -= _Q(i,j);
_Q(j,j) -= _Q(j,i);
}
}
}
double average_rate = 0.0;
for (unsigned i = 0; i < nstates; i++)
average_rate -= pi[i]*_Q(i,i);
double scaling_factor = 3.0/average_rate;
_Q *= scaling_factor;
// S is a symmetric matrix
eigenMatrixXd_t S = eigenMatrixXd_t(_sqrtPi*_Q*_sqrtPiInv);
// Can use efficient eigensystem solver because S is symmetric
Eigen::SelfAdjointEigenSolver<Eigen::MatrixXd> solver(S);
if (solver.info() != Eigen::Success) {
throw XStrom("Error in the calculation of eigenvectors and eigenvalues of the codon model rate matrix");
}
_eigenvectors = _sqrtPiInv*solver.eigenvectors();
_inverse_eigenvectors = solver.eigenvectors().transpose()*_sqrtPi;
_eigenvalues = solver.eigenvalues();
}
The element i,j of the instantaneous rate matrix in this codon model is 0 if codon states i and j differ by more than 1 nucleotide. The line
_Q = Eigen::MatrixXd::Zero(nstates,nstates);
initializes all elements of the rate matrix _Q to zero, so we only have to change those cells in which codons i and j are identical or differ by just one nucleotide. If the two codons differ by one nucleotide, and if the amino acid coded by both codons is identical, then this is a synonymous change and the rate is 1 times the frequency of codon j (the codon resulting from the change). If the “from” and “to” codons differ by just one nucleotide and the amino acid coded by each differs, then this is a nonsynonymous change and the rate would be omega times the frequency of codon j (the “to” codon). If codons i and j are identical, then the rate is the negative sum of all other rates in the same row.
As was done for the nucleotide version, the rate matrix is calibrated so that one unit of time equals one nucleotide substitution per site. This calibration allows us to treat edge lengths (in units of substitutions per site) as if they were time. The calibration involves calculating the average rate implied by the matrix before calibration and then dividing each element of the rate matrix by scaling_factor, which is this average rate. We must also multiply by the factor 3 to account for the fact that each codon state is 3 nucleotides long.