Here we explore the problem of underflow when computing likelihoods of large trees, and how to deal with this in BeagleLib.
As trees get larger, the likelihood for an individual site becomes smaller, and, if the tree is large enough, the site likelihood will become, at least for some sites, too small to represent even as a double precision floating point number. You might suppose that this is not a problem because site likelihoods are stored as log(site likelihood) and are thus immune to underflow; however, due to the fact that unknown ancestral states must be integrated out, the site likelihood must first be computed before its log can be taken.
There is an easy way to handle this: rescale the conditional likelihoods (or partials) at several points during the postorder traversal that culminates in the site likelihood so that, at each of these points, the largest partial likelihood for any given site is 1.0. For example, if the likelihood for a site conditional on an internal node having states A, C, G, and T is, respectively, 0.49, 0.01, 0.01, 0.01, divide all 4 values by the largest, 0.49, to yield 1.0, 0.0204, 0.0204, 0.0204. This procedure ensures that the most important partial will never become too small, and it is of no consequence if, in the process, the other partials become negligible relative to that largest value because in that case they would have no influence on the site likelihood anyway. The site likelihood calculated in this way will of course be incorrect due to these rescalings, but can be corrected by multiplying the final site likelihood by all of the scaling factors that were removed along the way. In practice, the sum of the log of all scaling factors removed is added to the log of the (scaled) site likelihood.
A simple 4-taxon, 1-site example serves to illustrate the process, even though rescaling would never be necessary for a tree so small. Assuming the JC69 model and all edge lengths equal to 0.383119218 results in all transition probabilities equal to either 0.7 (if the state at the beginning and end of an edge are identical) or 0.1 (if the states are different), which makes hand calculation much easier. Further assume that, at the site in focus, each of the four taxa has state A. The partials and final site likelihood calculation for this case without rescaling are shown below.
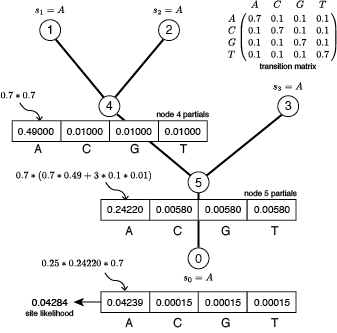
Now consider factoring out a scaler at both node 4 and node 5.
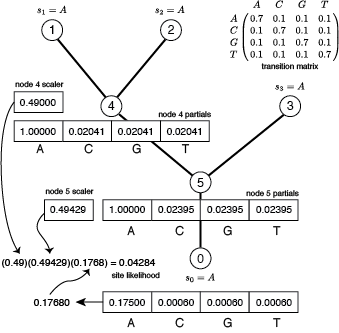
As you can see, the site likelihood is the same, however using scalers we avoid underflow. Above, the site likelihood is shown as a product of 3 numbers: 2 scaling factors and the rescaled site likelihood. Taking the log of this product to obtain the log site likelihood would undo all the work that went into rescaling. In practice, BeagleLib adds the log of each scaling factor to a cumulative sum as soon as they are calculated, and once the rescaled site likelihood is computed (0.1768 in the example above), its log is added to the cumulative sum of log scaling factors to obtain the log site likelihood. Performing the final multiplication on the log scale is what allows us to avoid underflow.
| Step | Title | Description |
| Step 12.1 | Large Tree Likelihood Fail | An illustration of underflow in calculation the likelihood of a large tree. |
| Step 12.2 | Rescaling in BeagleLib | Set up rescaling in BeagleLib to deal with underflow. |